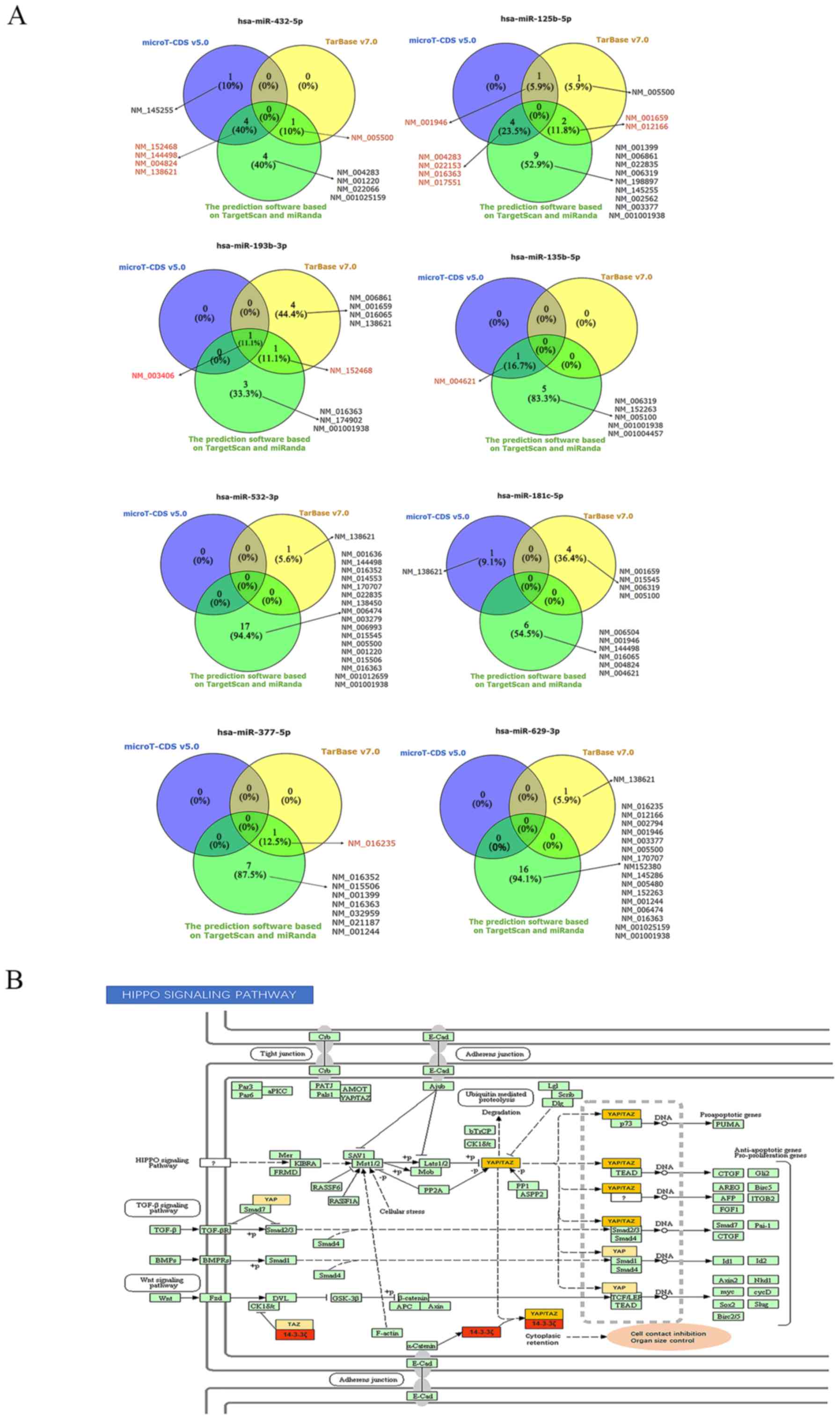
Interplay between SARS‐CoV‐2‐derived miRNAs, immune system, vitamin D pathway and respiratory system - Karimi - 2021 - Journal of Cellular and Molecular Medicine - Wiley Online Library
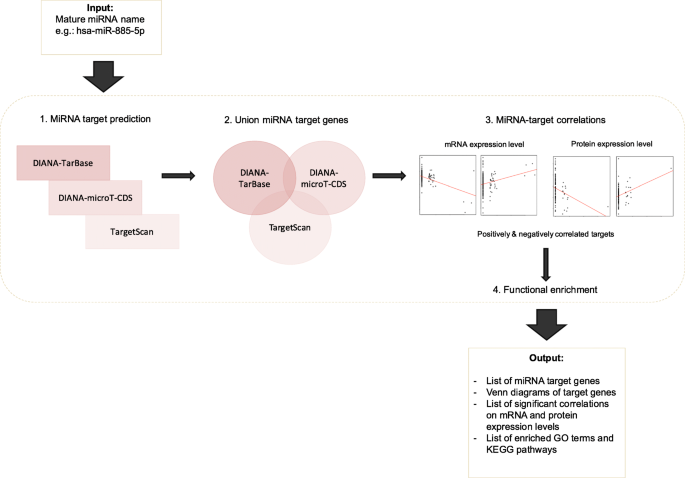
miRFA: an automated pipeline for microRNA functional analysis with correlation support from TCGA and TCPA expression data in pancreatic cancer | BMC Bioinformatics | Full Text
miR-125b and miR-532-3p predict the efficiency of rituximab-mediated lymphodepletion in chronic lymphocytic leukemia patients. A French Innovative Leukemia Organization study | Haematologica

Target prediction program comparison. Venn diagrams comparing predicted... | Download Scientific Diagram

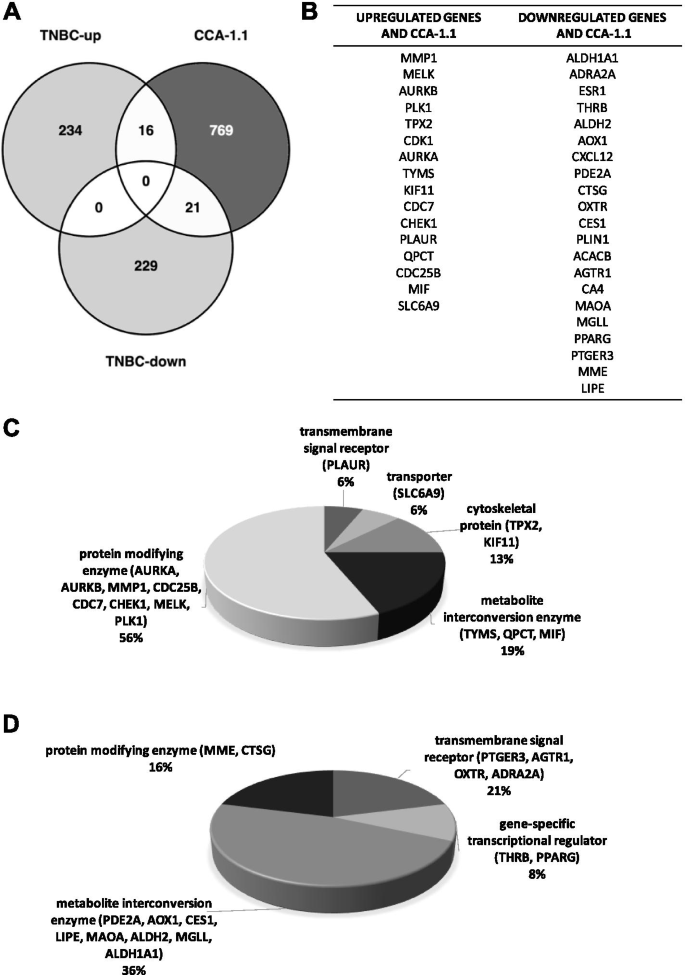
Predicting Target Genes of San-Huang-Chai-Zhu Formula in Treating ANIT-Induced Acute Intrahepatic Cholestasis Rat Model via Bioinformatics Analysis Combined with Experimental Validation

Target gene prediction and candidate gene determination. a Venn diagram... | Download Scientific Diagram
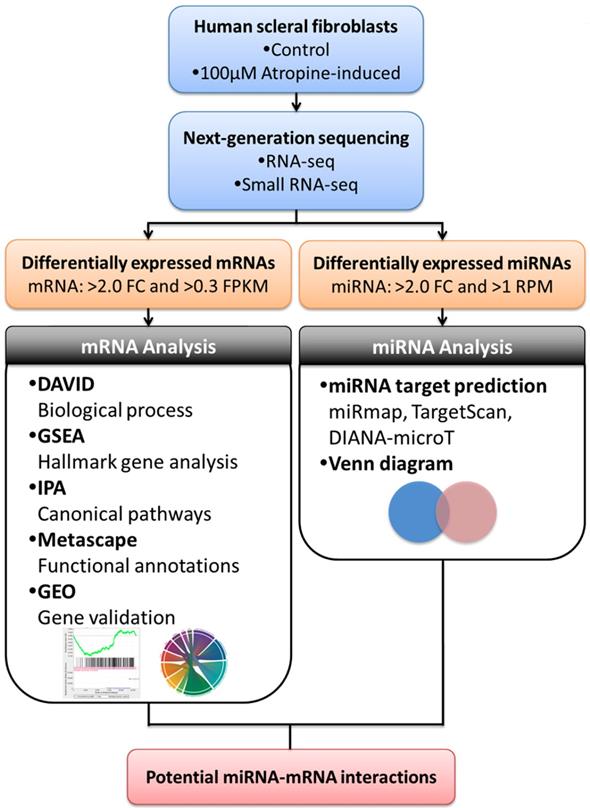
Systematic Analysis of Transcriptomic Profile of the Effects of Low Dose Atropine Treatment on Scleral Fibroblasts using Next-Generation Sequencing and Bioinformatics
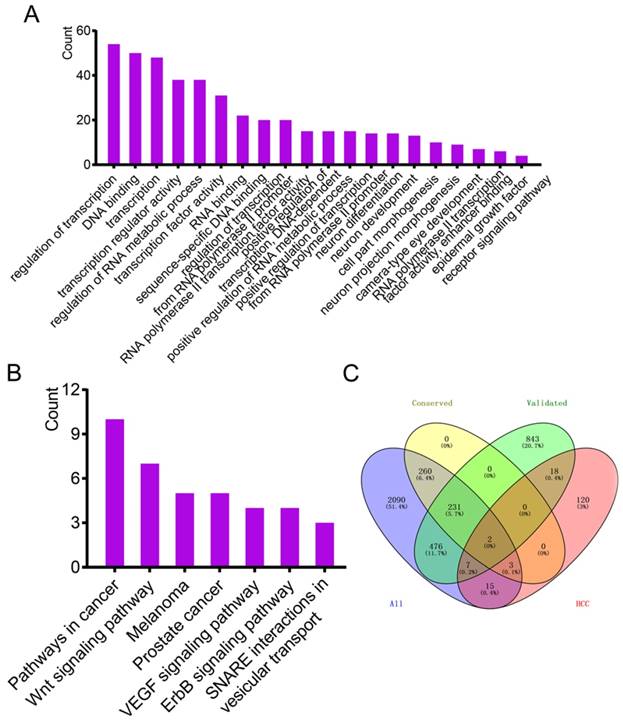
Clustered microRNAs hsa-miR-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for hepatocellular carcinoma

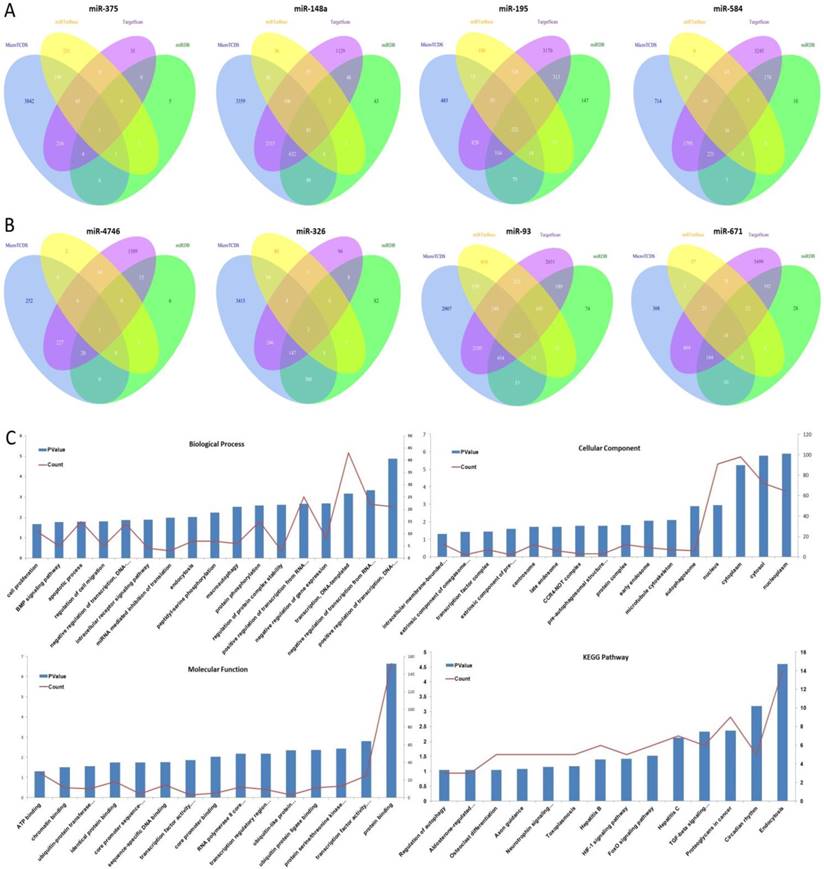
Integrative Bioinformatics Analysis Identifies Noninvasive miRNA Biomarkers for Lung Cancer | medRxiv
PLOS ONE: Identification of Keratinocyte Growth Factor as a Target of microRNA-155 in Lung Fibroblasts: Implication in Epithelial-Mesenchymal Interactions

Genes | Free Full-Text | Identification of Long Non-Coding RNAs Involved in Porcine Fat Deposition Using Two High-Throughput Sequencing Methods | HTML
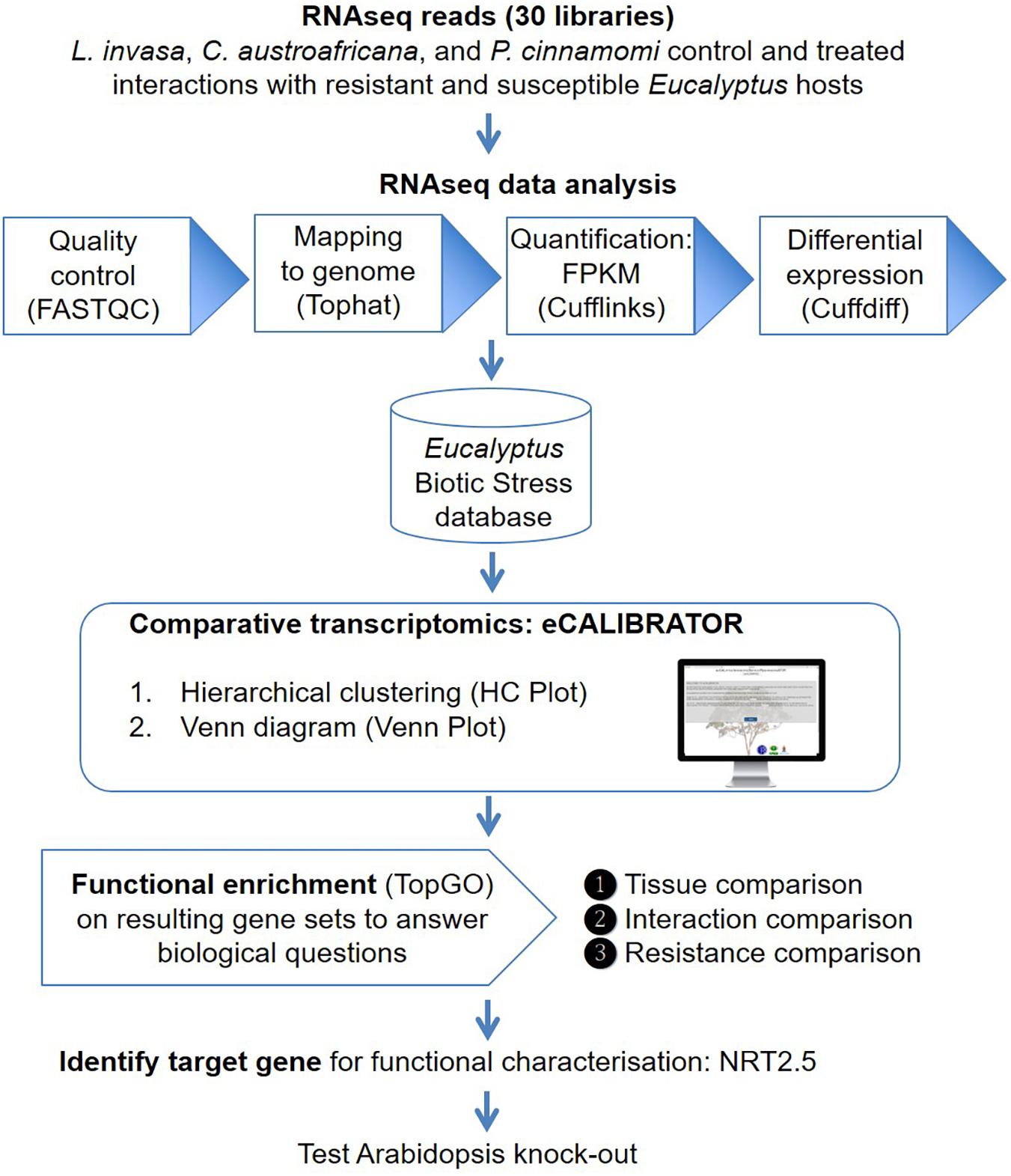
Frontiers | eCALIBRATOR: A Comparative Tool to Identify Key Genes and Pathways for Eucalyptus Defense Against Biotic Stressors | Microbiology
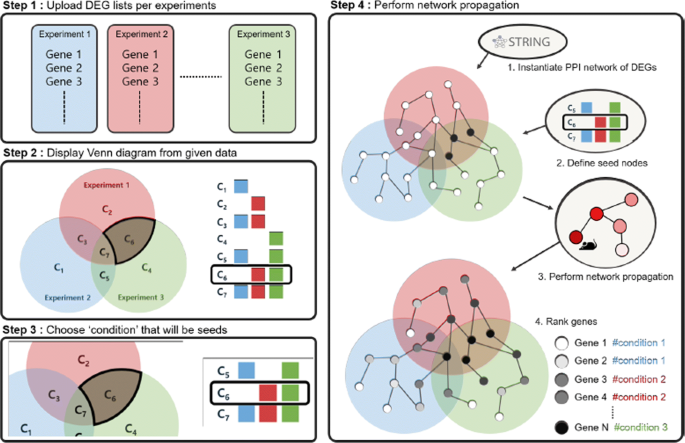
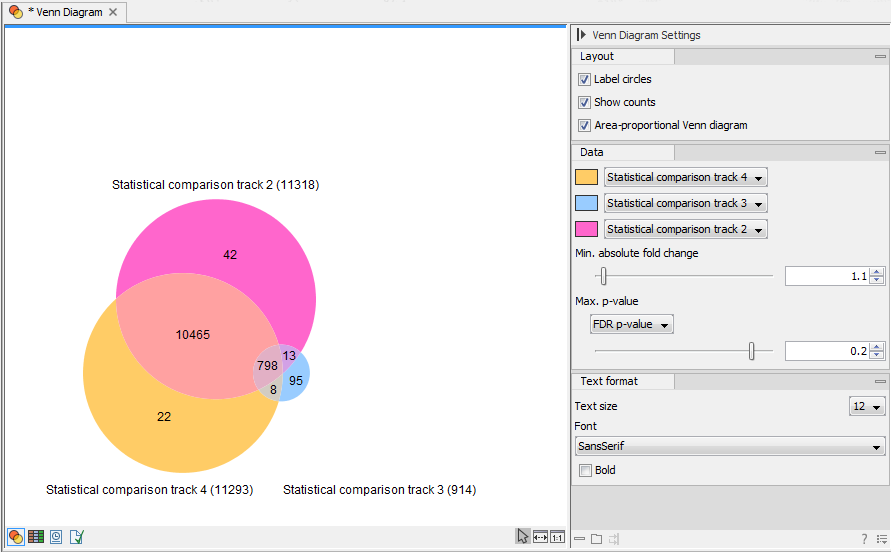
Venn-diaNet : venn diagram based network propagation analysis framework for comparing multiple biological experiments | BMC Bioinformatics | Full Text

Prioritizing target-disease associations with novel safety and efficacy scoring methods | Scientific Reports

miRNA-93 Inhibits GLUT4 and Is Overexpressed in Adipose Tissue of Polycystic Ovary Syndrome Patients and Women With Insulin Resistance | Diabetes